Week 9: Wrapping Things Up!
Monday August 26, 2019
Happy Monday! This past weekend was so relaxing! It was kind of hard to get up for work today haha. And I faced a bit of a surprise when I arrived at the museum because my badge didn’t work anymore! I was supposed to end the internship last week, but we extended it for one week and we didn’t let Access Control know… oopsies
It’s all fixed now, don’t worry :D
After that mini roadblock, I continued to examine the models and make changes to improve them. In my model training to find the differences between Lycopodium and Selaginella, some folds did really well and training accuracy reached 90% while validation accuracy lagged behind, but in other folds, the training and validation accuracy both plummeted to around 50% by the 10-15 epoch and just couldn’t recoveer. In all epochs, however, loss was constantly decreasing. I’m not really sure what’s happening there and why it would happen on some folds and not others… maybe there’s a few images that are just really throwing the model off? Although I did look through them manually before. Perhaps it’s worth another look.
I first looked into doing image augmentation, but I originally thought that it would result in the model training on more images than you have every epoch. Turns out, it just trains on a smaller sample, but each epoch it uses randomly altered images that are different to the model. It may be useful in another application (like the frullania), but currently when we have so many images already, I don’t think we need it.
I tried playing around with the regularizers today in the Dense and Convolutional-2D (Conv2D) layers, but most combinations I tried resulted in the model overfitting very early and the training and validation accuracy would begin to decrease by the 6-7th epoch. (To see what changes I made, check out this doc. The model started to learn again pretty well when I kept activity regularizers in the Dense layers, despite there not being an exact equivalent in the Smithsonian Methematica Model.
We’ll see what happens tomorrow morning!
-Al
PS, Matt wants me to try running the model again on the two frullania species again but the images being from farther away. I want to look into bootstrapping for this and hopefully synthetically expand our data set.
Tuesday, August 28, 2019
First thing this morning, I checked on the training of the model on Lycopodium and Selaginella. For the most part, it was okay; there was still a bit of overfitting, but the trend of validation accuracy seemed to follow training accuracy, like in the graph below.
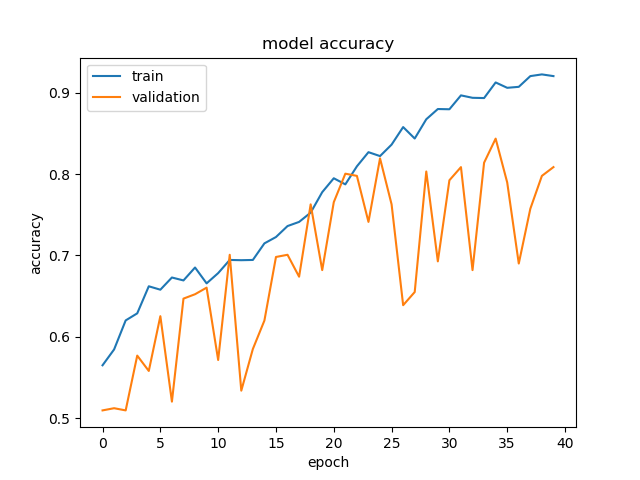
However, on other folds, we got something disastrous like this:
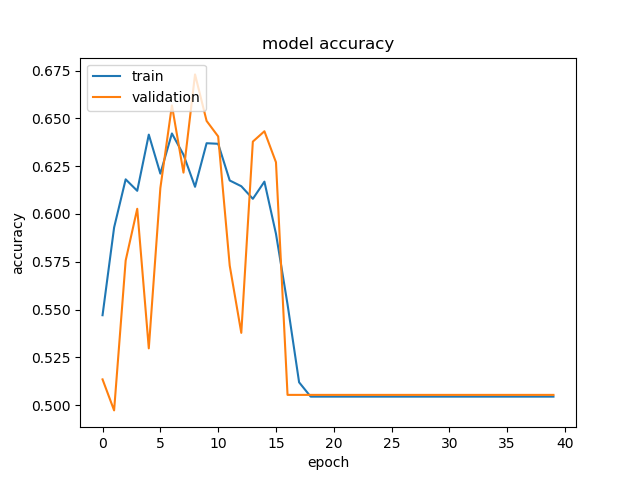
I’m not entirely sure why on most folds, the model seems to be training well, while on others, it’s completely unable to after a certain amount of time. I don’t think it’s overfitting because even the training accuracy decreased. Well, tomorrow, I’m meeting with Dr. Iacobelli, the computer science professor at Northeastern Illinois University, so hopefully he’ll be able to offer more insight.
Additionally, I received new images of the frullania species today. These ones are more zoomed in and hopefully will allow the model to pick up on more details in the differences between the two plants. I implemented some bootstrapping to train the model because we only had a little less than 300 images which I’m not sure is enough to be able to completely train the model on. I left it going overnight, so I’ll be able to check in on Thursday.
At this point, most of my big machine learning changes are wrapping up and I’ve been working on documentation, documentation, and documentation! Especially since I was pretty much the only one working on this project this summer, it’s extremely important that someone could study my work for a bit and pick up where I left off.
Thanks for reading!
-Al
Wednesday August 28, 2019
This morning I met with Dr. Iacobelli, Beth, and Dr. Campbell (unfortunately Matt coudln’t make it) We updated Dr. Iacobelli and here are his responses/advice. Regarding the random dropped accuracy I wrote about yesterday, he recommends that we meticulously check the code first. There may a bug (such as divide by 0) that’s throwing everything off. Additionally, it simply may be lack of processing power in the computer causing a data overflow.
Another thing he noticed is that the images we’re using from the Field Musuem are not consistent in the herbarium sheet layout. Some have labels in the upper right corner, some are lower left, some have specimen in the middle, some have specimen in the lower right corner. The inconsistencies in layout may be providing too much noise for the model, so he recommends that we try a smaller sample of images that have a more consistent layout an see if there is an improvement in performance.
Additionally, there is a computer at NEIU with 64 GB RAM that Beth is going to be trying our model with the Smithsonian images (which I didn’t know we had!) and we found out there is a computer at the museum with one terabyte of RAM. I repeat, of RAM. I almost fainted when I heard that.
Although I’m wrapping up this week, I’m excited for Beth to be able to try things out!
-Al
Thursday August 29, 2019
When I came in this morning, the model that was training on images of the Adiantum and Blechnum genera finished training, so I created a script to test it. Since the model really consists of 10 trained models (given that we used 10 fold cross validation), I had to take that into account. One way to do this would be see which prediction output each model gave and the final prediction would be what the majority of the models output. However, this poses two possible issues:
- What if 5 models say class A and 5 say class B? There wouldn’t be an odd number in this case to be the tie breaker
- If one model is 99% sure that an image is class A but another model is only 50% sure that it’s class A, using the method described above would give each of these equal weights when it maybe shouldn’t.
To overcome this, I decided to take a step back and look at the probability outcomes that are used to determine each model’s final answer. Basically, the final layer of the model outputs two numbers: the probability that an image is one class or another. So what I did was I kept track of each of these probabilities and for each image, added up all the models’ class A probability and class B probability separately. The final answer would be whichever summed probability is higher. This way, the “confidence” of each of the 10 models is taken into account.
Honestly, that didn’t take me very long today, so when I tested the Adiantum/Blechnum model, we got a whooping 97.8% accuracy!! It was tested on 400 new images and this is the best I’ve ever gotten :)))
This is good news because it means the model can work! We just need to tweak values and perhaps look at cleaning our input images for Lycopodium and Selaginella.
The rest of the day was mainly spent documenting my work. I explained to Matt how my Google Drive was organized and how to use Github as well as documented within the Drive where everything is.
I did, however, forget to upload my final cross-validation testing file to Github, which is what I’ll do first thing tomorrow morning.
Last day tomorrow! What a melancholy feeling…
-Al
Friday August 30, 2019
Today was spent mainly double checking my documentation on Github and Google Drive. I called Beth and we went over each file in Github and she seems confident on where I’m leaving off, which is great! I’m so excited for her to continue this project and be able to run the model on a computer that has enough processing power.
Furthermore, Matt worked on getting the images contrasted a bit more yesterday; now the background of the images are very white compared to the specimen in focus and it helped get rid of some noise. We only have a little less than 150 of either group, but we ran the model for 50 epochs and we actually got some results! Although overfitting still occurred, the validation accuracy definitely grew as the number of epochs increased! This is a great sign! It means the machine is able to generalize what it’s learning on the training data onto new images.
Since we didn’t run for very long and we don’t have many images, we were able to start another run before the end of the day. Matt went through the images and deleted any that were actually the wrong specimen or weren’t sterile as this type of noise could be hurting the model’s performance. Then, before I left, I ran two models: one with the same exact specs as the last run, just with the new images, and one with the new images and more epochs. Although I won’t be able to see the results, Matt will be able to check it out and report back!
And that’s a wrap :) although I’m leaving, I know this project is going to go far in the hands of Beth, Matt, and Dr. Iacobelli. I’m excited to see where they are able to take this application of computer science into botany and am truly grateful for this opportunity!
-Al