Week 8: Implementing Cross Validation
Monday August 19, 2019
This morning, Beth came to the Field Museum to work with me on implementing stratified cross validation and ROC testing. Stratified cross validation essentially partitions the data into groups of approximately equal distributions of each class. ROC (stand for Receiving Operator Curve) is a type of graph that can help us quantify how “good” our model is. It uses the percentage of true positives and false positives and the closer the curve hugs the y-axis, the more confident it is. Take a look at the image below:
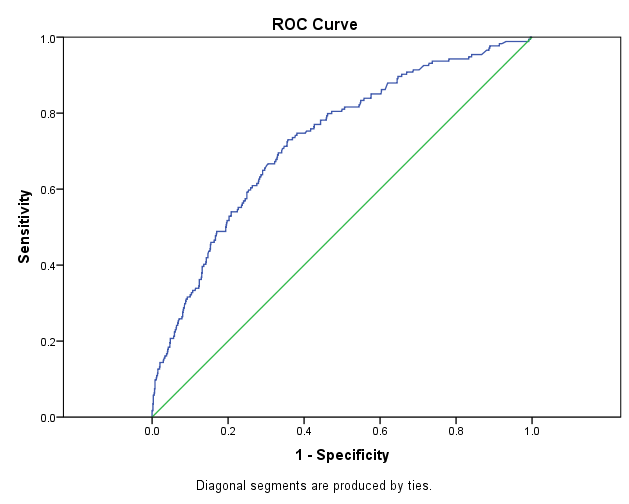
The straight line represents if it were just up to chance, so if the ROC curve is close to that, it’s bad.
I got the code to run with stratified cross validation and creating an ROC curve at the end with small amounts of bogus test data, but when I tried to import larger amounts of useful data, the program seems to be crashing and I’m not sure why. I ran the old model again (prior to implementing cross validation) and it seemed to work okay, but in this new script, it feels like everything’s falling apart (rip). Before the program started crashing, it would train but ridiculously overfit on the training data while validation accuracy didn’t increase at all :(
I’m trying to see if I accidentally changed something when adapting the model, but then the code crashed entirely, freezing the computer.
This is definitely a rough note to end the day on, but I’m going to try not to let it bother me too much and come in tomorrow ready to problem solve!
-Al
Tuesday August 20, 2019
This morning, I was able to fix why the code would crash my computer! When I was using the split function on the stratifiedKFold object and accessing features and labels that made up the training and validation sets, they had to be numpy arrays. My labels data structure was only a list and thus would throw an error and crash it. After making a few more tweaks on random seeds and epochs, aroudn 10:30 AM I started running the 10 fold cross validation and it hasn’t stopped since. I’ve left it on and hope no one messes with it! Tomorrow morning, we’ll be able to check out results and see what changes we can make to improve the model.
While the model was training, I was tasked with a side task. There’s a bryophyte checklist of the Java region that’s written as a word document. We want to parse the document and store the genus, species, and founder into a CSV. Currently, I’m using a package called python-docx and basic python string manipulation to parse out the desired information. The only downside is this code I’m writing is only specific to this document which of course isn’t as fun in CS.
Have a great day!
-Al
Wednesday August 21, 2019
Today was a long day, oof. To start out, Matt and I talked about the project and thought about why our results weren’t exactly matching that of the Smithsonian. First, and foremost, it’s most likely because of our data that we’re using. The Smithsonian is using images from their own herbarium while we’re using images from the Field Museum herbarium and obviously those will produce different results. Within the family of Lycopodiaceae, the distribution of images across genera are different between their herbarium and ours. Additionally, they were able to use a lot more images due to processing power. I found out today, that using around 4000 images is a lot for the computers we have (i5 Intel Cores with 8 GB RAM). Sometimes the computer just freezes entirely and you have to hard restart it. Thus, I decreased my number of images to around 3300 and it seems to be running okay!
Additionally, instead of comparing families of plants (Lycopodiaceae and Selaginellaceae), we decided to simplify the inputs to two genera of plants (Lycopodium and Selaginella). Selaginella is the only genus in the family Selaginellaceae (as a reminder the hierarchy goes family > genus > species) and Lycopodium is one of around 8 or so genera in the family Lycopodiaceae. Hopefully this will provide less noise in the first class and make it easier for the machine to detect prominent differences.
Additionally, just for kicks, Matt wanted me to run the model to compare two new and different genera: adiantum and blechnum. This is because these two families look VERY different to the eye, so the model should do a relatively good job with this. I spent most of today preparing the images to be run on a different computer and started running the model before I left.
I got in around 8:15 this morning and didn’t leave till around 5:30, I’m going to go sleep a lot now!
-Al
Thursday August 22, 2019
I came in this morning and good news, nothing crashed! The models are still chugging along doing their own thing. Something I noticed and continue to notice is that while training accuracy will rise somewhat consistently with each epoch, validation accuracy fluctuates a LOT more, like in the graph below:

I’m going to look into this more, but after a quick Google search, it seems one issue may be too much dropout. So after this model is finished training, I’m going to look into making that change (which might end up having to be done tomorrow)!
The program I started running yesterday with Lycopodium and Selaginella was taking forever… after 18 hours, it only completed 5 folds of the 10 fold cross validation and Beth and I decided that it was too long to realistically experiment. Thus, we decreased the images from 256 x 256 pixels down to 128 x 128 and it seems to be going much faster! Each epoch now takes a little over a minute instead of around 10 minutes.
Additionally, I’m running a baseline test with the model and images of Frullania rostrata and Frullania coastal. As a reminder, these are two possibly distinct species that we want to gather further evidence that they are different. Although the model isn’t “tuned” to picking up the differences in the Frullania, it’ll be nice to have a baseline essentially and see how to go from there. Beth had also mentioned that there’s a way to see what features each layer of the model is picking up, which would be helpful in our case to see what layers are working for our purpose and what should be changed.
Take it easy!
-Al
Friday August 23, 2019
Happy Friday! When I came in this morning, all three of my models finished running and here’s a brief summary of the results:
The Lycopodium/Selaginella results: The validation accuracy followed the training accuracy, but there was still a lot of fluctuation. In my next training, I increased batch size from 32 to 64 and increased epochs to 100. May as well since we have the weekend to run it. Before I left for the day, it finished training 2/10 folds and I noticed in the second fold, by the end neither training nor validation accuracy increased from 50%. This leads me to believe we shouldn’t increase batch size much above 32.
The Adiantum/Blechnum results: This run was mostly as a baseline to see whichout changing the model, what results did we get. Again, they were pretty similar to before: validation accuracy seemed to follow training accuracy but with a LOT more and larger fluctuations. In this next training session, I decreased the initial dropout layer from 50% to 40% to see if perhaps less measures against overfitting would help.
The Frullania (coastal/rostrata) results: awful. I really didn’t expect much but what we saw here in all the folds was a classic case of overfitting. The training accuracy kept increasing while validation accuracy remained fluctuating around 50%. To make this work, we would need to reconfigure the layers of the model to “detect” these certain features.
On a more fun note, today our department had a large rpotluck! There were all sorts of food from Malaysian, Latin American, to Chinese! I brought carrots and hummus and a Chinese Eight Treasures Cake which got really good feedback! I ate literally so much food but it was absolutely amazing :)
Have a great weekend in this beautiful weather!
-Al