Week 3: Statistical Analysis for Microplants, Image Editing, and Basic Machine Learning
Monday July 8, 2019
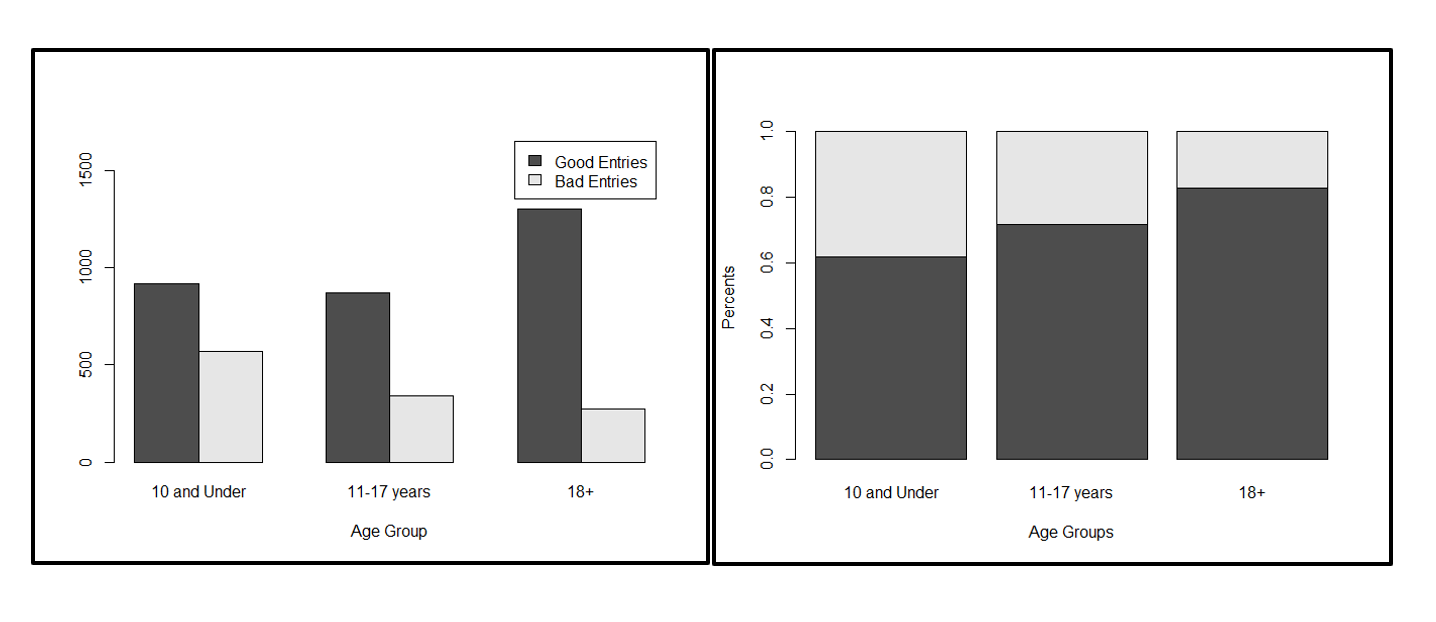
Hope everyone had a great July 4th weekend! Today, we were back on the grind, starting with analysis of the Microplants data that we previously cleaned. To jog your memory, recall that the purpose of us cleaning Microplants kiosk data and combining it with age data is to see if there is a difference in the accuracy of measurements (determined by the angle between the major and minor axis) between the different age groups. Today, I worked on learning how to use R and without even doing any statistical tests and just graphing the data, we can see that there in fact is a difference between groups, as shown below.
The next task here is performing a linear regression model and chi square test to have a statistical measurement of how different the results are depending on age group and how much more likely one age group is to perform more accurate measurements than another age group.
Additionally, today I played around with GIMP, an image editing software that is used to binarize the images of specimen for MorphoSnake to read in. I was able to eliminate the background of an image, but I’m still working on getting only the desired parts of the specimen black. The tricky part here is that we don’t want the lobules (leaf looking like parts) to overlap, since that could mess up the morphosnake measurements. The good news is that in most of the images you can see both dorsal and ventral lobules (where ventral lobules show the front and dorsal show the back) The ventral lobules tend to be more “skeletal” and if we can just keep those, that’s sufficient. Currently, we’re working on creating a script that will be able to recognize which pixels are part of the stem and ventral lobules and be able to discard the dorsal parts, but it’s very much in the process.
Thanks for reading!
-Al
Tuesday July 9, 2019
This morning was spent working on the statistical analysis of our Microplants data in R. We’re performing a binomial test on the data (since our results have two options: good or bad) and initially I was facing issues with the data structures in R. Then I learned that matrices in R are homogenous, meaning they can only hold data of one type, while data frames are heterogeneous, so they can have both strings and integers. After implementing those changes, we were able to get the glm (general linear model) function working with our data and we found that the data is statistically significant, since the probability (p-values) were incredibly low. Additionally, we calculated the odds ratio which help us determine just how much more likely one group is to create a good measurement than another.
After working on the statistics side in the morning, we had a team meeting with the rest of botany, the paleobotany department, and the Digital Learning Internship students. We did another round of introductions, which is helping me learn everyone’s names! Additionally, Jose and I explained our projects and did a quick demo of MorphoSnake to show them how the software worked. Our explanation was pretty quick, but I hope it made sense!
After lunch, I found this plug-in for GIMP (the image processing software) that allows you to write python scripts using GIMP, but I’m struggling to get it working on Ubuntu. First, there aren’t as many resources out there regarding this area and a lot of the tutorials assume things that aren’t there in our version of Ubuntu and GIMP, so a lot of it is trying new things and working from there. At the same time Jose is working on just using Python packages to do the image editing and hopefully between the two of us exploring different paths, we’ll be able to get something going soon.
Today was tiring but pretty rewarding, get some rest!
-Al
Wednesday July 10, 2019
Okay not gonna lie today was not super productive, but it’s okay, we all have those days. In the morning, I went to the intern breakfast downstairs and got to meet some of the other interns at the museum! We also had a Q&A with the president of the museum and it was really cool to see this more personable side and actually kind of get to know him. After that, I was working more with GIMP and MorphoSnake, trying to get an image of a plant skeletonized. I was able to take the following image and kind of skeletonize it (Not sure if I could actually say I was successful) I ended up making it black and white, intensifying the contrast, using the smart scissors tool to select more of the stems, and eventually using the paint tool to draw out the stem. However, when I tried using it in MorphoSnake, it threw an exception. Still trying to figure out why it wasn’t accepting this image, but in the mean time, I started working on other things.


The right is an unedited image taken under a microscope and on the right is a skeletonized version.
After lunch, I started the TensorFlow Machine Learning package sent from Beth, a graduate student at Northeastern Illinois University. I basically followed a tutorial using keras and tensorflow to create an algorithm that matches pictures of articles of clothing to the correct name. It uses 60K training cases and 10K test cases. Although I don’t fully understand what each statement is doing, I understand the general gist of the program, and I think it’s a good place to start before I move onto more complex ML problems. Additionally, next week, Jose and I will be hosting a Python workshop, so I went through his notes and added some of my thoughts. I think learning computer science to teach others computer science is one of the most exciting parts of the field! It’s something that everyone wants to learn about these days, and I hope the other interns/people attending our quick class are excited to learn! Lastly, I started documenting and commenting in the Microplants cleaning script. I want someone to be able to look at the code and understand what every line does. Tomorrow, that’s something I’ll continue working on.
Happy Hump Day!
-Al
Thursday July 11, 2019
In the morning, I helped organize the herbarium, taking a break from all this computer stuff. Basically, the museum is in the process of digitizing the specimen collection, but there were a few fern specimen that have to be re-imaged or didn’t get imaged in the first place. Teaming up with some interns from the Digital Learning Internship, we looked for those sheets and organized them to be sent to the imaging lab. Although it’s not the most exciting work, it’s definitely necessary and it was nice to get to know some of the high school interns better!
In the afternoon, I worked on MorphoSnake stuff again. Using the paint tool in GIMP, I took an image of a plant, traced what I thought would be the stems and threw that image into MorphoSnake. Something that I learned is somethimes, after editing the image, MorphoSnake can’t always accept PNGs if they have alpha layers or transparencies. So to be safe, I started using JPGs. Below, you can see the different stages of this process. The only issue is that drawing them by hand is not necessarily something the machine can learn to do easily. Thus, we may need to look into a different method of skeletonizing the images.
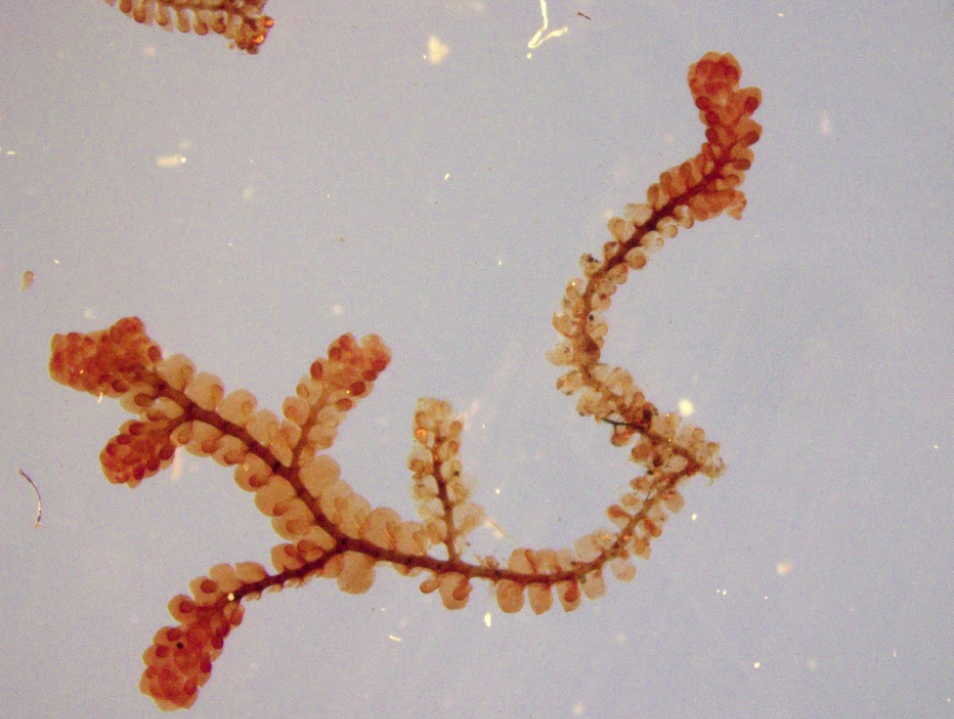

Left: unedited image of frullania rostrata. Center: Skeletonized image of f. rostrata using paint tool on GIMP. Right: Skeletonized image used in MorphoSnake.
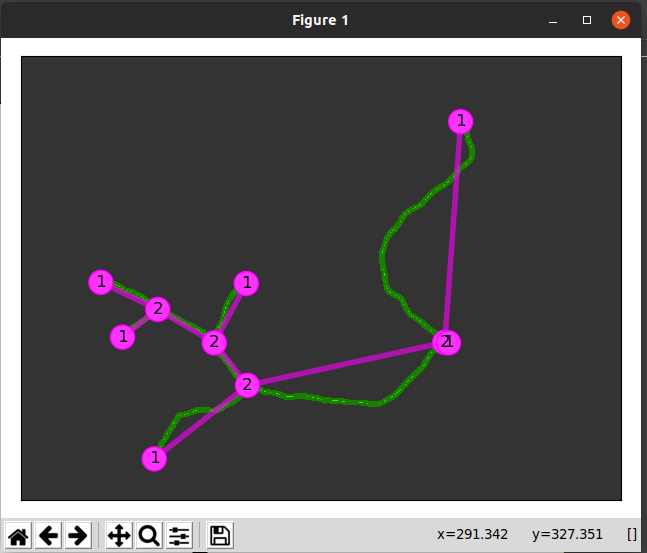
Since the technique above may not be reasonable to get a computer to do, I explored altering the image as a whole. For example, to obtain the sequence of images below, I took the following steps:
- Use the fuzzy select tool and set the threshold relatively high (around 80) to select entirely the main object)
- Go to Select -> Invert
- Choose the fill color to be white and use the bucket fill option to make the background all white.
- Go to Color -> Threshold and adjust the slider values until a somewhat desired image is produced. The histogram shows how many pixels of the image are in that range. Pixels inside the range are converted to white and everything else is black.


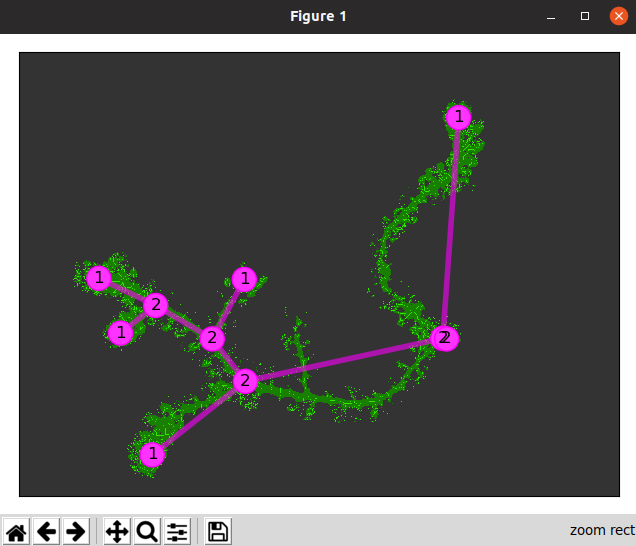
Left: unedited image of frullania rostrata. Center: Skeletonized image of f. rostrata using whole image editing on GIMP. Right: Skeletonized image used in MorphoSnake.
I’m currently trying to look into other options for image editing. Victoria suggested looking into Google Vision API, but that seems to be more just for recognition of objects in the image. It may be a place to start, but I’m not sure if it can do the image editing for us. I’ll have to check in with Jose tomorrow and touch base on ideas.
A bit of a block, but we’ll get through it!
-Al
Friday July 12, 2019
he morning was spent organizing with the high school interns again! Additionally, I spent some time looking more into convolutional neural networks and reading up on them to understand a bit more. I also looked into gradient descent functions to minimize cost, since those are pretty commonly used. In the afternoon, I worked with Jose to add some finishing touches to the Python workshop that we’re hoping on hosting next Thursday for the other interns/employees in botany, paleobotany, and the high school interns. On a completely other note, the next step in the Microplants project will be to match each measurement entry with the correct species. We have a list of subject ID’s and their corresponding species from someone who worked on this a year ago, so our next step will be to incorporate this matching into the cleaning function. I didn’t think of this originally, but we could look into implementing a hash table for easy look up. It’s been a while since I worked with those, so I’ll have to look into the purposes and syntax in Python again.
Short and sweet ending, enjoy your weekend!
-Al